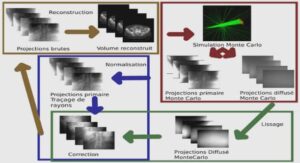
Ciblage thérapeutique au sein des cellules endothéliales du cerveau via l’utilisation d’immunoliposomes PEGuylées ciblant le récepteur de la transferrine
Materials and methods
mAbs radiolabeling
One mCi of N-succinimidyl-[2,3-3H] propionate ([3H]-NSP, 101 Ci/mmol) was evaporated under a nitrogen stream in a borosilicate tube and incubated with 500 mg of mAbs into 100 ml of a buffer containing 0.1 M sodium borate/1.5 M NaCl (pH 8.0) for 1 h at room temperature, followed by a 2 h incubation at 4oC. Radiolabeled mAbs were separated using a G-25 Sephadex gel filtration column (GE-Healthcare-Amersham, Baie d’Urfé, QC, Canada) pretreated with 10% (w/v) BSA (Sigma-Aldrich, Oakville, ON, Canada), and specific activity was computed using disintegrations per minute (dpm) counts and protein content in eluted fractions.
DNA labeling
DNA was labeled using the ULYSIS nucleic acid labeling kit (Alexa Fluor 546, AF546) following the manufacturer’s instructions (ThermoFisher/Life Technologies). Briefly, two generic plasmids were digested with restriction enzymes to generate equimolar linear DNA fragments of size < 1.1 Kb (1097, 1069, 1057, 1008, 896, 179 and 1008, 725, 610, 472, 467, 370, 105 bp) that were purified by ethanol precipitation. Then, 200 µg were resuspended in 1200 µl of labeling buffer, denatured at 95°C and kept on ice while 3 vials of ULS labeling reagent were prepared and pooled in 300 µl 50% dimethylformamide. Denatured DNA was then incubated with labeling reagent at 80°C for 45 min and unbound fluorophores as well as solvant were washed away with Montage columns (Millipore, Etobicoke, ON, Canada). Degree of labeling was calculated at one molecule of AF546 for each 38 bp of DNA and 200 µg in 200 µl H2O were added to the lipids. Water was used for encapsulation instead of ethanol.
PEGylated stealth immunoliposomes (PSILs) preparation
Briefly, POPC(1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine) (Northern Lipids Inc., Vancouver, BC, Canada) (18.6 mmol), DDAB (Didodecyldimethylammonium bromide) (Sigma-Aldrich) (0.6 mmol), DSPE-PEG2000 (distearoylphosphatidylethanolamine)-PEG2000 (2,000 Da polyethylene glycol) (Northern Lipids Inc.) (0.6 mmol) and the linker lipid DSPE-PEG2000-maleimide (0.2 mmol) (Nektar Therapeutics, Huntsville, AL) were dissolved in chloroform, followed by evaporation under a nitrogen stream and constant agitation in order to produce a thin lipid film, which was allowed to dry during 30 min. The dried lipid film was dispersed in 0.2 ml 0.05 M Tris-HCl (pH 7.0), vortexed for 2 min, followed by a 10 min incubation, 10 min of bath sonication and a second 1-min vortex agitation. Fluorescent dsRNA analog siGLO Red (Dharmacon, Ottawa, ON, Canada) (40 nmoles), or fluorescent DNA (200 µg) were added to the lipids. Then, 0.6 ml of a 67 % (v/v) ethanol solution in 0.05 M Tris-HCl buffer (pH 7.0) was added drop by drop during 30 sec under minimal agitation. Large vesicles were converted into liposomes under 100 nm by extrusion using a hand-held LiposoFasti-Basic extruder (Avestin, Ottawa, ON, Canada) as described (Shi, Y Zhang, et al. 2001; Yun Zhang, Calon, et al. 2003). mAbs were conjugated to the freshly formed liposomes by adding three mg of unlabeled mAbs (RI7 or 2a3; BioXcell West Lebanon, NH) and 0.5 mCi of tritiated mAbs were thiolated with a 40:1 molar excess of freshly prepared 2-iminothiolane (Traut’s reagent, Sigma-Aldrich) following a 1-h incubation at room temperature in 0.05 M sodium borate/0.1 mM EDTA (pH 8.5). Thiolated mAbs were transferred to a buffer containing 0.05 M HEPES/0.01 mM EDTA (pH 7.0) by centrifugation in a Vivaspin (Sartorius Stedim Biotech, Aubagne, France) filter unit (MWCO 30 kDa). This step was repeated twice and the final volume was reduced to ~200 µl. Afterwards, thiolated mAbs were added to the liposome preparation for an overnight room temperature incubation in a 2 ml glass bottle under inert nitrogen atmosphere. siGLO Red and fluorescent pDNA encapsulation and mAb conjugation efficiencies were determined by size exclusion chromatography (Figure 1). PSILs were eluted with 0.05 M HEPES (pH 7.0) at a rate of 1.0 ml/min in a 1.5 cm 20 cm pre-equilibrated Sepharose CL-4B column (GE-Healthcare-Amersham, Baie d’Urfé, QC, Canada). Fractions were quantified to determine their 3H and fluorescent content by liquid scintillation and fluorescence quantification using a Synergy HT (Biotek, Winooski, VT). Eluted fractions containing PSILs were kept in glass vials under nitrogen atmosphere and stored at 4oC unless specified otherwise.
In vitro uptake and colocalization analyses
Murine neuroblastoma (N2A, #CCL-131, ATCC, Manassas, VA) and murine brain endothelioma (bEnd5, #96091930, HPACC, Salisbury, UK) cells were seeded in 96-well plates containing DMEM supplemented with 10% FBS at 4 x 105. Two days later, medium was removed and cells were incubated with DMEM-10% FBS containing Ri7-PSIL containing siGLO Red (500 nM) for 15, 45, 60 min, 4 and 24 h. Afterwards, cells were washed with ice-cold DMEM-FBS 10%, 1X D-PBS and fixed with 4% paraformaldehyde (PFA) for 15 min. For colocalization analyses, following the fixation, cells were permeabilized with D-PBS containing 0.5% Triton X-100 for 10 min and blocked with a 3% bovine serum albumin (BSA), 3% normal horse serum (NHS) and 0.05% Triton X-100 during 45 min. Cells were then incubated overnight at 4oC with primary antibodies in a D-PBS solution with 1% BSA, NHS and Triton X-100: rabbit anti-EEA-1 (1:200, NEB Cell Signaling, ON, Canada), rabbit anti-TfR ab84036 (1:200, Abcam, ON, Canada) and rabbit anti-LAMP-1 (1:2000, Sigma-Aldrich). Following the incubation with primary antibodies, cells were exposed to Alexa-FluorTM-conjugated donkey anti-rabbit (ThermoFisher Scientific), cell nuclei were counterstained with DAPI (Sigma-Aldrich) and IBIDI mounting media was added. Images were acquired with an EVOSTM FL auto imaging system (ThermoFisher Scientific).
In vitro competition studies
N2A cells were seeded in 96-well plates containing DMEM supplemented with 10% FBS at 5 x 105. The next day, medium was removed and cells were preincubated with Ri7 or control IgG (2a3) (0, 50 or 200 nM) for 30 min. Then, Ri7-PSILs were added to a final siGLO Red concentration of 250 nM and incubation was resumed for 15 min. Incubation was terminated by sequential washes with ice-cold DMEM-FBS 10% and D-PBS 1X. Afterward, cells were fixed with 4% PFA, cell nuclei were counterstained with DAPI (Sigma-Aldrich) and IBIDI mounting media was added. Images were acquired with an EVOSTM FL auto imaging system.
Animals
Adult Tie2GFP mice (Jackson Laboratory, Bar Harbor, ME) weighting 20-30 g were used. They had ad libitum access to food and water. All procedures were performed in accordance with the Canadian Council on Animal Care standards and were approved by the Animal Ethics Committee of the Centre Hospitalier de l’Université Laval (CHUL).
Tissue preparation for post-mortem analysis
For microscopy experiments mice were systemically injected with Ri7- or 2a3-PSILs containing 20 µg (1.33 mg/kg) of siGLO Red, pDNA546 and sacrificed 1 h post injection by terminal transcardiac perfusion under deep anesthesia with ketamine/xylazine. Animals were perfused with 25 ml of cold 1X PBS followed by 50 ml of 4% PFA, pH 7.4. Brains were rapidly dissected, post-fixed with 4% PFA for 4 h and cut in coronal sections of 25 µm with a microtome (Leica Microsystems, Richmond Hill, ON, Canada). For TfR levels quantification animals were injected twice a day for two days and sacrificed by transcardiac perfusion with 1X PBS containing protease inhibitors (SigmaFast protease inhibitor tablets, Sigma-Aldrich) 24 h following the last injection. Brain and liver were collected and weighed.
Brain microvessels isolation
Mouse brain microvessels were isolated by density-gradient centrifugation using the capillary depletion technique as previously described (Alata et al. 2015; Do et al. 2014). Briefly, brains were collected and transferred into ice-cold 1X PBS. Cerebellum, meninges and brain stem were removed. Afterward, brains were gently homogenized in ice-cold DMEM containing 10% FBS using a Potter homogenizer. The homogenates were centrifuged at 500 xg for 10 minutes at 4°C, the supernatant was excluded and the pellets were homogenized in 5 ml of ice-cold DMEM containing 25% BSA and centrifuged at 1,500 xg for 20 minutes at 4°C. The resulting pellets were suspended in 1 ml of ice-cold DMEM with 10% FBS and the homogenate was filtered through a 60-µm filter. The filtrates were centrifuged at 12,000 xg for 45 minutes at 4°C. The pellets containing the microvessels were washed in ice-cold 1X PBS and centrifuged again at 12,000 xg for 20 minutes at 4°C. The supernatants were discarded and the pellets were stored at – 80°C until processed for western blotting analysis.
Proteins extraction
Proteins were extracted from tissues as previoulsy described (St-Amour et al. 2014). Briefly, the pellet containing the microvessels was weighed and total proteins were extracted by homogenization in four volumes of lysis buffer (150 mmol/L NaCl, 10mmol/L NaH2PO4, 1% Triton X-100, 0.5% SDS, and 0.5% deoxycholate, pH 7.4) containing Complete protease inhibitors (Roche, Indianapolis, IN, USA) and 10 mg/ml pepstatin A (Sigma-Aldrich). The obtained suspension was sonicated briefly (3×10 seconds) and centrifuged at 100,000 xg for 20 minutes at 4°C. Supernatant was stored at − 80°C until western blotting analysis.
Western Blotting
Equal amounts of proteins per sample were added to Laemmli’s loading buffer, heated to 95°C for 5 minutes before loading (20 µg protein per lane), and subjected to sodium dodecyl sulfate-polyacrylamide (SDS) gel electrophoresis. Proteins were electroblotted onto PVDF membranes (Immobilon, Millipore, MA, USA) before blocking in 5% non-fat dry milk, 0.5% BSA, and 0.1% Tween-20 in 1X PBS for 1hour at room temperature. The membranes were washed three times for 10 minutes in 1X PBS containing 0.1% Tween-20. Then, membranes were incubated overnight at 4°C with rabbit anti-TfR primary antibody (1:1000, Abcam) diluted in 1X PBS containing 0.1% Tween-20, 5% non-fat dry milk, and 0.5% BSA. The next day, membranes were washed three times for 10 minutes in 1X PBS containing 0.1% Tween-20 and then incubated for 1 hour at room temperature with goat anti-rabbit horseradish peroxidase-labeled (1:60 000, Jackson, West Grove, PA, USA) secondary antibody diluted in 1X PBS containing 0.1% Tween-20 and 1% BSA. The membranes were again washed three times for 10 minutes in 1X PBS containing 0.1% Tween-20 and probed with chemiluminescence reagents (Luminata, ThermoFisher Scientific). Immunoblots were analyzed with a KODAK Imaging Station 4000MM Digital Imaging System (Molecular Imaging Software version 4.0.5f7, Carestream Health, Rochester, NY, USA).
Immunofluorescence
Washing steps were performed using 1X PBS, pH 7.4, between every step of the immunofluorescence protocol. Coronal brain sections were blocked for 1 h with a 1X PBS solution containing 5% NHS (ThermoFisher Scientific) and 0.2% Triton X-100. Afterwards, sections were incubated overnight at 4oC with primary antibodies in the blocking solution: mouse anti-neuronal nuclei (1:1000, NeuN, Chemicon/Milipore, Temecula, CA) and mouse anti-GFAP (1:1000, Sigma-Aldrich). Following incubation with primary antibodies, slices were exposed to Alexa Fluor conjugated donkey anti-mouse and anti-rabbit secondary antibodies (1:1000, ThermoFisher Scientific). Finally, slides were then coverslipped with ProLong® Diamond antifade media (ThermoFisher Scientific).
Results
Uptake studies of Ri7-PSILs were performed on mouse N2A neuroblastoma cells and bEnd5 murine endothelial cells, which were incubated with Ri7- or 2a3-PSIL containing a fluorescent dsRNA analog (siGLO Red) for 15, 45, 60 min, 4 and 24 h (Figure 34). Uptake of the Ri7-PSIL complex was observed within 15 min and increasing over a 24 h time period, while no signal was visible in cells incubated with 2a3-PSIL (Figure 34). Pretreatment with 50 or 200 nm of free Ri7 blocked the accumulation of Ri7-PSIL in N2A to a level similar to the control nanoparticle (Figure 35A-D). In comparison, pretreatment with 2a3 mAb did not alter the uptake of Ri7-PSIL (Figure 35E). Finally, colocalization analyses showed a robust colocalization between TfR and Ri7-PSIL (Figure 35F). These results confirm the ability of Ri7-PSIL to ferry nucleic acids into cultured N2A or bEnd5 cells, via the TfR.
To investigate the cellular fate of Ri7-PSILs, we incubated N2A cells with the anti-TfR nanocomplex containing siGLO Red for 15 min, 1 or 4 h and performed colocalization experiments with intracellular markers by immunofluorescence. The results of the immunofluorescence with EEA-1 showed that after 1 and 4 h of incubation on N2A cells, Ri7-PSIL had enter endosomal compartments (Figure 36). However, colocalization between Ri7-PSIL and EEA-1 was barely visible after 15 min, suggesting a slow internalization process. Moreover, the cellular distribution of the fluorescent signal suggests that most fluorescence from siGLO Red in Ri7-PSIL was still located at the cellular membrane after 15 min (Figure 36).
To access whether Ri7-PSIL were directed toward lysosomal compartments, we performed colocalization experiments with N2A cells incubated with Ri7-PSIL for 15 min, 1 or 4 h and lysosomal marker LAMP-1 (Figure 37). Colocalization analyses at 15 min showed that Ri7-PSIL were not rapidly directed toward the lysosome. Colocalization between siGLO signal and LAMP-1 immunolabeling was hardly visible after 15 min, but suggest that Ri7-PSIL slowly accumulated into lysosomal compartments after 1 and 4 h. However, our data suggest only a partial accumulation after a 4 h incubation on N2A cells (Figure 37C).
To assess the brain targeting capacity of Ri7-PSIL, Tie2GFP mice were injected with Ri7- or 2a3-PSIL containing fluorescent pDNA (Figure 38) or dsRNA siGLO Red (Figure 39). Animals were sacrificed 1 h after the systemic administration. Data in Figure 38 show fluorescent pDNA encapsulated in Ri7-PSIL was extensively distributed into the cerebral microvasculature, colocalizing with with GFP signal from the Tie2GFP endothelial cells (Figure 38A-C). No signal was detectable in the brain of mice injected with fluorescent pDNA complexed with PSIL conjugated with the control IgG 2a3 (Figure 38D). Further experiments were conducted with Ri7-PSIL containing a dsRNA fluorescent analog (siGLO Red) (Figure 39). Similarly, clear signal was observed in the GFP-labeled cerebral microvasculature in animal injected with siGLO-Ri7-PSIL formulation, but not when PSIL were conjugated with a control antibody. Overall, using fluorescent pDNA (Figure 38) and siGLO Red (Figure 39) presented data show that Ri7-PSIL delivered genetic material to BCECs in the mouse in vivo. In all experiments conducted, no signal from the nanocomplex targeting the TfR was visible outside the brain vasculature.
Previous study using anti-TfR antibodies suggested that lysosomal accumulation could lead to both in vitro and in vivo TfR degradation and thus impaired the uptake of anti-TfR mAbs administered chronically (Bien-Ly et al. 2014). To investigate if Ri7-PSIL could be used for repeated administration without altering TfR levels, Tie2GFP mice were injected twice a day for two days with Ri7-PSIL. Quantification by Western blot on capillary depletion fractions showed no significant difference on TfR levels between mice injected with Ri7-PSIL and control uninjected mice (Figure 40). Thus, our data suggest that even though Ri7-PSIL tends to accumulate toward lysosomal compartments in vitro, TfR levels remained unchanged in vivo after repeated injection of anti-TfR nanoparticles.