Facteurs de variation des effets de Nosema ceranae chez l’abeille une approche de la virulence
Obtaining N. ceranae spores for experimental infection
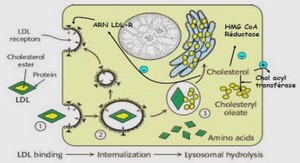
Bee samples from both geographic locations obtained as described above, were processed in parallel, in exactly the same way, to obtain fresh mature spores as described in Botías et al. (2011). Briefly, the abdomens of all bees were homogenized in 25 ml H2O PCR grade for 2 min at high speed in a Stomacher® 80 Biomaster (Seward, West Sussex, UK) using strainer bags (BA6040/STR, Seward). The homogenate was recovered in a tube and 15 ml of H2O PCR grade was again added to the strainer bag to repeat the homogenization under the same conditions. Honey bee homogenates were centrifuged (6 min at 800g), the supernatant was discarded as the spores remained in the sediment. The pellet was resuspended in 1 ml of distilled water. To confirm the Nosema species of the spores, an aliquot of each homogenate was analyzed by PCR as previously described (Martín-Hernández et al., 2011b) using 218MITOC FOR/218MITOC REV and 321APIS FOR/321APIS REV primers specific for N. ceranae or N. apis respectively and COI-F/COI-R primers for A. mellifera COI, as internal control of each reaction. All the PCRs reactions were carried out in a Mastercycler® ep gradient S (Eppendorf®, Hamburg, Germany). Each PCR product was analyzed in a QIAxcel System (Qiagen, Hilden, Germany), using a QIAxcel DNA High Resolution Kit (Qiagen, No. 929002) to detect positive and negative reactions. Negative controls were analyzed in parallel to detect possible contaminations in all phases of this technique. Once the N. ceranae species were confirmed, the spores were purified with Percoll® to obtain fresh pure spores suspension for artificial infection. During extraction and before infection, spores were kept at room temperature. The spore number was counted using a hemocytometer chamber and a phase contrast microscope
Genetic variability of an rDNA fragment from two N. ceranae isolates
Up until now no reliable genetic markers have been found to be suitable to distinguish between strains of N. ceranae. In fact, two genetic markers can produce different phylogenetic trees of the same sample, leading to incongruous results (Ironside, 2007). Because of this, we performed a study on the variability of an rDNA fragment between samples as in Sagastume et al. (2011), through the amplification by PCR and cloning of the product, to look for a decrease in variability that consequently occurs because of geographic isolation. Total DNA was previously extracted from honey bee samples naturally infected by N. ceranae from Central Spain and South of France (see above). PCR was performed with the pair of primers named NOS3-UPPER (5´ACTGGCTTAACTTCGGAGAG 3´) and NOS3-LOWER (5´AAGTAATACCGTTACCCGTCA 3´) which amplify a 890 bp fragment that contains the intergenic spacer (IGS) and part of the small subunit (SSU) of rDNA. In order to ensure the reliability of our results, PCR was performed using a low initial DNA concentration and a high fidelity polymerase with proof-reading capability that works with an elongation time of one minute, enough to synthesize 890 bp in each cycle of PCR (Meyerhans et al., 1990; Bradley and Hillis, 1997). PCR reactions started with 5 µL per tube of 1/10 diluted DNA, 0.4 µM of each nucleotide, 0.4 µM of each primer, 1.5 units of Expand High Fidelity Plus PCR System (Roche, cat no. 3300226) and its 5X buffer, 12 µg of BSA (Roche Diagnostic, cat no. 10711454001) per tube and sterilized distilled water for a final volume of 25 µl. PCR was performed in a Eppendorf Mastercycler EpGradient Pro S thermocycler with the following program: 94°C for 2 min followed by 35 cycles of 94°C for 30 sec, 57.2°C for 30 sec, and 68°C for 1 min, plus a final step at 68°C for 7 min. The PCR products were kept at 4ºC, and 5 µL of each one were resolved on standard 2% agarose gels (Invitrogen E-GEL 2% Agarose GP, cat no. G8008-02) and visualized by ethidium bromide staining. 4 µL of each PCR product were cloned in Escherichia coli plasmid pCR2.1-TOPO® with TOPO TA Cloning® Kit (Invitrogen, cat no. K4500-01) following the manufacturer’s instructions. From 4 to 6 clones were obtained per PCR product and the plasmid DNA was extracted and purified using the QIAprep Spin Miniprep Kit (Qiagen, cat no. 27106). A sample of each plasmid was digested with EcoRI (New England biolabs, R0101S) and separated in 1% agarose gel electrophoresis in order to check the correct size of the insert. The inserts from the different clones were sequenced, using the commercial primers M13, at the Alcalá de Henares University, Unidad de Biología Molecular of the Faculty of Environmental Sciences, Spain. Fourteen sequences from GenBank (http://www.ncbi.nlm.nih.gov/nucleotide) were used to compare our results (Table 1). All the sequences were aligned following the CLUSTAL W algorithm Thompson et al., 1994) with the program BIOEDIT 7.0.5.2 (Hall, 1999), and the analysis of polymorphic sites and haplotypes generation were carried out with the program DNASP (Rozas et al., 2003)
Experimental infection
Newly emerged honey bees of the local subspecies A. m. iberiensis were obtained as described in Higes et al. (2007) and Martín-Hernández et al. (2009) from a pool of three N. ceranae and N. apisfree honey bee colonies with queens naturally mated. Briefly, frames of sealed brood were kept in an incubator at 34ºC ± 1ºC, and when new worker bees started to emerge, they were carefully removed and confined to cages in an incubator for five days at 33ºC ± 1ºC and fed ad libitum with sugar syrup composed of 50% sucrose solution and 2% Promotor L (Calier Lab) through an individual feeder attached to the cage. Three experimental groups of honey bees were assigned as follow: honey bees infected with N. ceranae from Avignon (South of France), honey bees infected with N. ceranae from Marchamalo (Central Spain) and a group of non-infected honey bees as control. Each group was composed of three replicates of 40 honey bees, 30 of them destined to follow mortality and the other 10 selected at random for optic and electronic microscopy preparations. The five-day-old bees were starved for 2 hours and then anesthetized with CO2 to be fed individually using a micropipette with 1 µl of a spore solution containing 40 000 spores per bee. After infection honey bees were fed ad libitum with the same sugar syrup from the beginning. Two similar incubators (Memmert® Mod. IPP500, ± 0.1ºC) were maintained at 33º C, one contained the N. ceranae infected honey bees and the other the uninfected honey bees.
Histopathological study
Two honey bees per cage were collected at random both at days 12 and 16 post-infection to be used for histology preparations as described in Higes et al. (2007) and Martín-Hernández et al. (2009). After dissecting out the alimentary canal, the ventriculus and attached Malpighian tubes were divided into sections and fixed in 10% buffered formalin for 24 hours. The tissues obtained were then embedded in paraffin, and 4 µm thick sections were stained with haematoxylin-eosin to perform a complete histopathological study as described in Higes et al. (2007). For transmission electron microscopy (TEM) we utilized the method described in Higes et al. (2007) and Martín-Hernández et al. (2009). From each replicate cage the ventriculus and attached Malpighian tubes of two honey bees were processed. Tissue was prefixed in a 2% glutaraldehyde – 2.5% paraformaldehyde solution for a maximum of one hour, before it was washed three times in phosphate buffer (PBS, pH 7.4) and postfixed in 1% osmium tetroxide at room temperature. After again washing in PBS and dehydrating with an ascending series of acetone, the ventriculus was embedded in Epon-Araldite resin. Semi-thin (0.5 µm) sections were cut with a Reichert-Joung ultracut E microtome, stained with 1% methylene blue in 4 % sodium borate water, and observed with an Olympus Vanox AHB53 photomicroscope. After light microscopy selection of representative tissue areas, the Epon block was trimmed before ultrathin (60 nm) sections were obtained. For TEM studies, grids were double contrasted with 2% uranyl-acetate in water and lead citrate Reynolds solution for 10 min each, then examined and photographed with a Jeol 1010 electron microscope at an accelerating voltage of 80-100 kV.